When Bacteria Outsmart Antibiotics, Surveillance Can Help Battle Resistance

The problem of so-called “superbugs” has emerged as a serious public health issue, with bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) or carbapenem-resistant Enteriobacteriaceae (CRE) among the pathogens that cause infections resistant to multiple antibiotics.
But what is antimicrobial resistance (AMR) and how do bacteria become resistant to antibiotics?
How Microbes Can Outsmart Antibiotics
The machinery inside of a bacterial cell makes the bacteria less susceptible to an antibiotic, and this can lead to resistance — so what is that machinery doing?
Since bacteria grow and reproduce quickly, they also quickly adapt on a genetic level to pressures from the outside through genetic mutations and even through acquiring genes from other bacteria. If those genetic changes make the bacteria resistant to a given antibiotic, then it will survive when it’s exposed to the antibiotic while others are killed off. In a short period of time, the resistant bacteria will multiply, passing the resistance to the next generations, and fill the space or infection site previously occupied by the less resistant bacteria. This process of selection — essentially one type of an evolutionary arms race between bacteria — describes how the development of antimicrobial resistance might occur.
The exact way that arms race plays out — the exact “contest” that takes place between bacteria — depends in part on the antibiotic used. For example, some antibiotics target a particular component of bacteria. The bacteria that are able to modify that component — the ones that win the “moving target” contest — will be the ones to survive and become more resistant.
Other forms of resistance that bacteria develop may be in the form of developing or acquiring an enzyme that inactivates the antibiotic, through modifying membrane transporter proteins so that a bacteria will no longer transport a particular antibiotic into itself, or by using proteins called efflux pumps to actually “pump” out, or extrude, the antibiotic. In all of these cases, the bacteria that are the best at performing these resistance activities will outcompete less resistant bacteria, and pass their resistance onto the next generations. Since bacteria reproduce so quickly, the ones that survive this selection process will proliferate and will be resistant to the antibiotic that killed off its competitors.
“Bacteria are, in a sense, clever,” says Jill Inverso, Pfizer’s VP of Critical Medical Care in the Collegeville, Penn., site. “Any of these mechanisms can come into play when bacteria are exposed to antibiotics.” But ultimately, she says, “it varies based on the pathogen and on the antibiotic.”
The Importance of Surveillance
At least 2 million people in the United States become infected with bacteria that are resistant to antibiotics, and at least 23,000 people die each year as a direct result of these infections, according to the U.S. Centers for Disease Control and Prevention.
The need for these kinds of strategies was underscored again in an alarming January 2017 report in which the CDC detailed the case of a Nevada woman who died after getting a CRE infection that was resistant to all 26 antibiotics available in the U.S.
“Bacteria develop resistance to every drug they’re exposed to, so we need a way to understand those trends on a broad scale,” Inverso says. “At the same time, we need to have specificity — by knowing which pathogens are problematic in which locations. Not all bacteria react to the pressure of antibiotic agents in the same way.”
For this reason, surveillance is a key step in the fight against AMR. By keeping tabs on antimicrobial resistance, surveillance can help doctors choose the most appropriate antibiotic against a particular pathogen. It can also help public health officials develop strategies to mitigate antimicrobial resistance.
One such surveillance system is the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) — a collaboration between local, state and federal agencies in the U.S. to track antimicrobial resistance in ill people, retail meats and food animals. NARMS can help public health officials monitor emerging trends, according to the CDC, which oversees the program along with the U.S. Food and Drug Administration and the U.S. Department of Agriculture.
In 2015, the World Health Organization launched an antimicrobial surveillance initiative called the Global Antimicrobial Resistance Surveillance System (GLASS), which collects and reports data on AMR rates aggregated at national level.
There is also a surveillance database called ATLAS, or Antimicrobial Testing Leadership and Surveillance, created by Pfizer 13 years ago at the request of U.S and European regulators. A new interactive website for ATLAS was launched in April 2017, allowing easy access to the database for doctors, scientists and public health officials. ATLAS tracks numerous pathogens and the extent to which each of them are resistant to dozens of antibiotics in more than 60 countries, and is updated every 6 months using data reported by various health institutions around the globe.
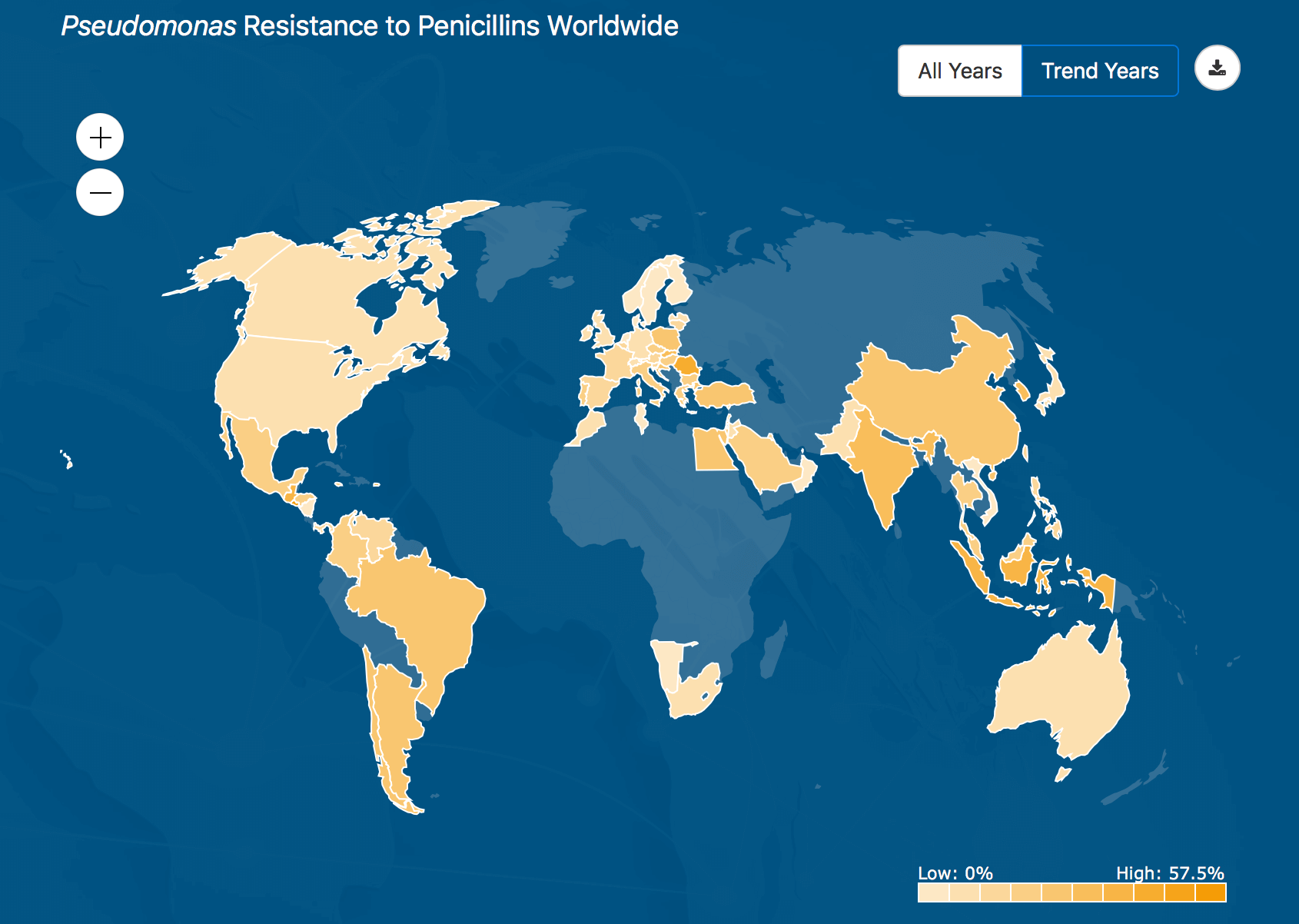
This heatmap shows where the Pseudomonas bacteria is most resistant to penicillin around the world, based on data collected by the ATLAS database from health agencies in 60 countries.
ATLAS is fully searchable, providing data on specific pathogens and antibiotics, including minimal inhibitory concentrations (MIC), the minimum concentration of an antibiotic needed to stop visible growth of a given pathogen. The new website also features a “heatmap” that allows users to choose combinations of bacteria and antibiotics and display the resistance rate. By moving the cursor, a pop-up box also displays the resistance rate for a specific country.
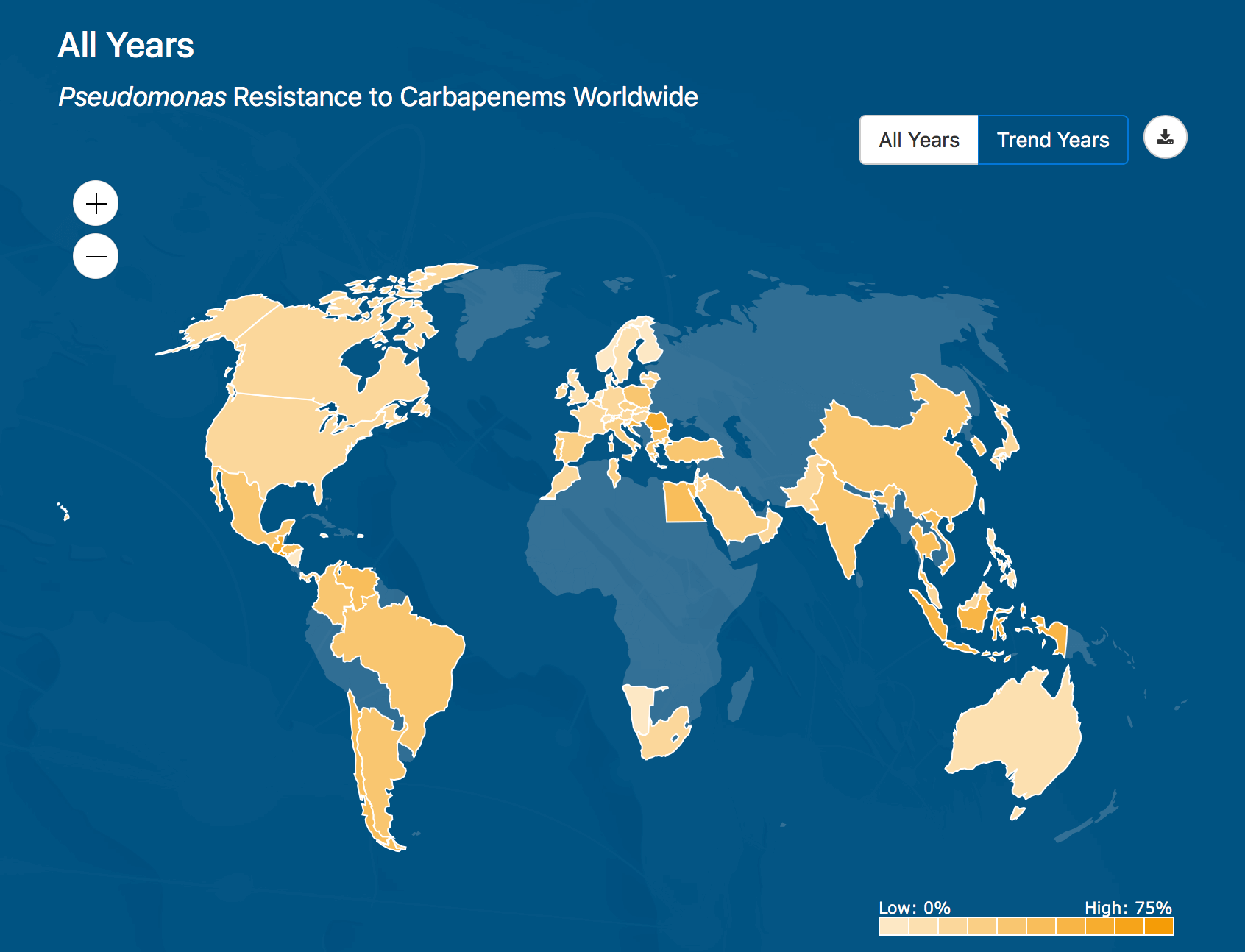
This heatmap shows where the Pseudomonas bacteria is most resistant to Carapenems around the world, based on data collected by the ATLAS database from health agencies in 60 countries.
“Until ATLAS, resistance wasn’t monitored in many parts of the globe, except for the U.S. and Europe,” Inverso says. “This is a huge step in making a well-informed fight against antimicrobial resistance.”