Using DNA ‘Barcodes’ in the Search for New Drugs

Organic lettuce, endangered sharks, and now, new medicines.
In recent years, DNA ‘barcoding’ technology has exploded as a tool to quickly identify and track plants, animals and the tiniest of molecules. Just as barcodes are used to identify products at the grocery checkout, DNA ‘barcodes’ are small bits of DNA code that can be used to quickly recognize and monitor materials.
In pharmaceutical labs around the world, scientists are using DNA-encoded compound libraries to screen billions of compounds, offering an efficient way to help find a molecular “hit," the starting point for developing drugs.
The technology is particularly promising for protein disease targets that have been historically difficult to work with. “The advantage of doing the DNA-encoded library screening is that you can screen many more compounds and cover much more chemical space than if you were screening a couple of million compounds in our standard, high-throughput screen methods,” says Adam Gilbert, a Senior Director in Medicinal Chemistry at Pfizer’s Groton, Connecticut research site. “You’re increasing the likelihood that you’re going to find a hit.”
A billion compounds in single test tube
Until recently, one standard method for finding new medicines has been high-throughput screening (HTS). Researchers isolate a target, such as an enzyme or receptor, or design experiments to assess changes in an entire disease pathway, and use these to screen against millions of liquid compounds. To run an HTS, scientists place each compound in its own “well,” (a tiny divot in a test plate with multiple “wells”) with a sample of the target protein to see how they interact. The process can be time-consuming and costly. Essentially, screening a million compounds can require running a million different experiments.
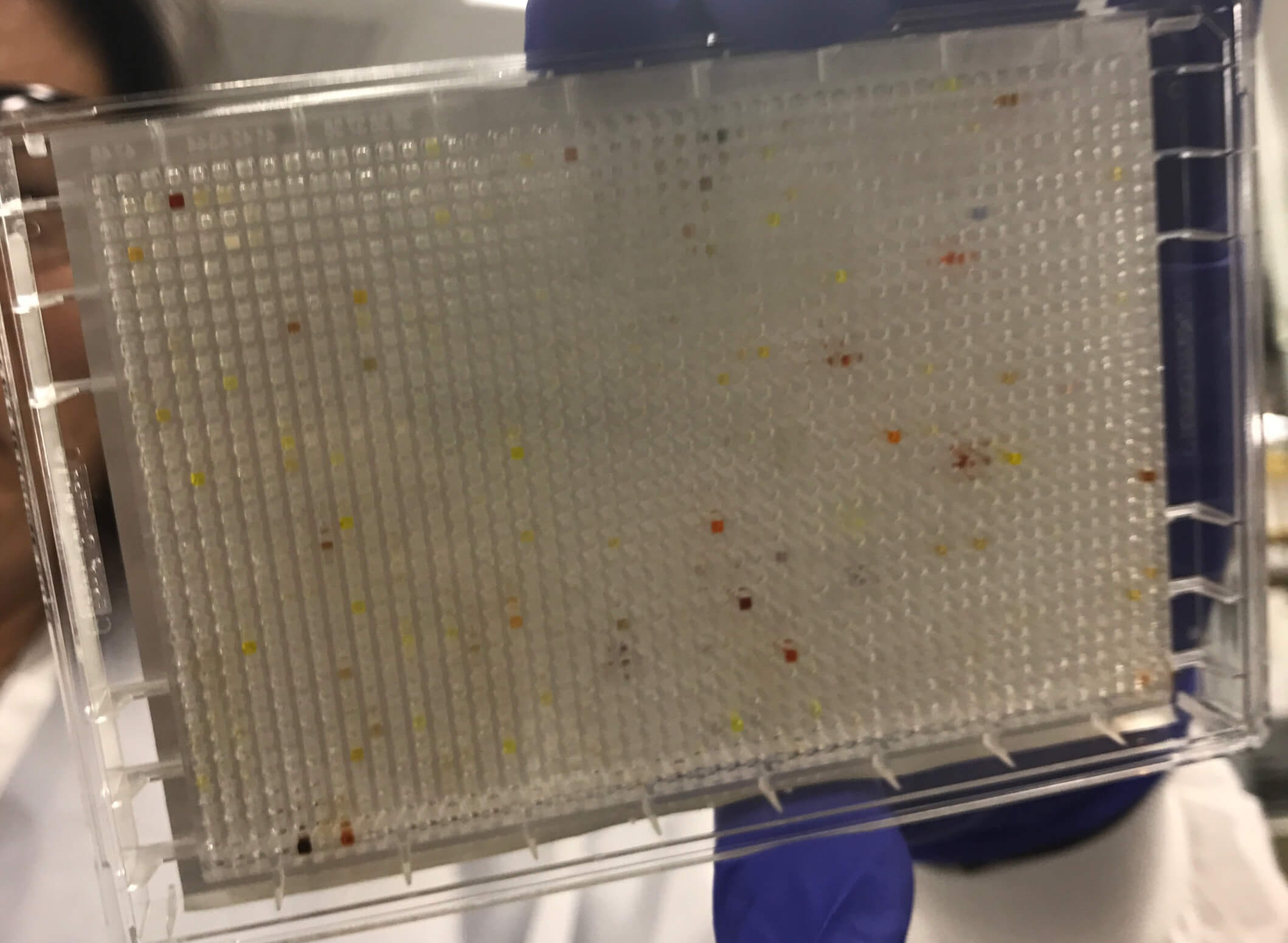
A cell assay plate with 1,536 wells.
Now, with the new technology that can serve as a complement to HTS, scientists can screen billions of chemicals at once in a single test tube. Scientists add the DNA-encoded compounds to a mixture with the target protein and see which ones “stick” and wash away those that don’t bind. Then, using gene-sequencing technology they read the DNA “barcodes” to see which compounds remain.
Going after the challenging targets
This technology will prove helpful in the coming years as scientists make new discoveries in oncology, immunology and rare diseases. The complex disease targets in these areas, such as certain cell receptors and protein-protein interfaces, have very large surfaces and shallow binding sites, so it’s difficult to find the right type of molecule that binds well to elicit biology. “Think of it like sticking your hand in a shallow, wide vase and trying to make a good fit with your hand. It’s hard if not impossible to do,” says Gilbert.
It’s also useful in cases where scientists have a very new target and want to do a quick initial test to see if it’s actually “druggable," meaning that a chemical structure can bind with it. “We can test billions of compounds very rapidly and cost effectively to see if anything sticks or not, and by the end of that process you have a good idea whether it’s going to be a protein that we can find binders for,” says Marie-Claire Peakman, Executive Director, Hit Discovery & Lead Profiling, Medicine Design, also based in Pfizer’s Groton site. DNA-encoded libraries also allow the flexibility to work with target proteins in multiple scenarios in parallel that could more closely resemble different cellular compartments or different stages of the protein life-cycle. “With HTS the cost and labor required means you can generally screen in only one set of conditions, but this new technology allows us to screen in multiple conditions in parallel, for example, with and without other protein binding partners or cofactors,” says Peakman.
But DNA-encoded libraries have their limitations as well. These initial screening tests can only reveal if a molecule will bind to a protein – they don’t show if it creates any functional activity, such as switching on or off receptor signalling. “That’s the biggest reason it won’t replace high-throughput screening. It can serve as complement to the process.” says Gilbert. “Ultimately, we’re looking for a compound that creates some functional activity. Therefore we will still require some downstream assays that can confirm whether the hits have the activity that we are hunting.”